iacs-ipac-reader
A reader plugin for read iacs/ipac images and export .rtdc files.
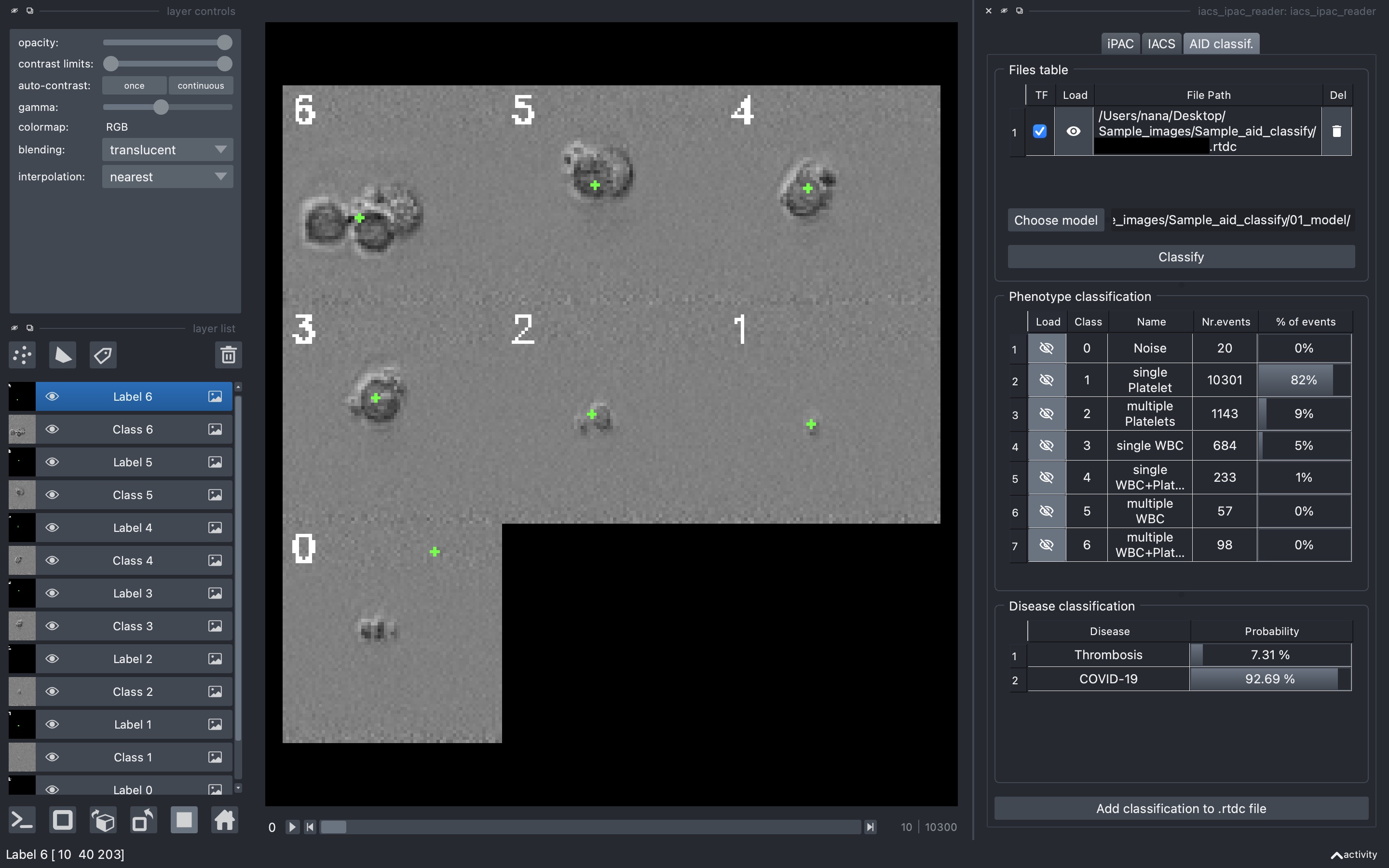
A plugin used a convolutional neural network (CNN) to distinguish single platelets, platelet clusters, and white blood cells and performed classical image analysis for each subpopulation individually. Based on the derived single-cell features for each population, a Random Forest (RF) model was trained and used to classify COVID-19 associated thrombosis and non-COVID-19 associated thrombosis.
More information about IACS/iPAC.
IACS: DOI: 10.1016/j.cell.2018.08.028
iPAC: DOI: 10.7554/eLife.52938
This napari plugin was generated with Cookiecutter using @napari's cookiecutter-napari-plugin template.
Installation
You can install iacs_ipac_reader via pip:
pip install iacs_ipac_reader
To install latest development version :
pip install git+https://github.com/zcqwh/iacs_ipac_reader.git
Introduction
The iacs-ipac-reader plugin mainly include 3 functional tabs:
- iPAC
- IACS
- AID classif.
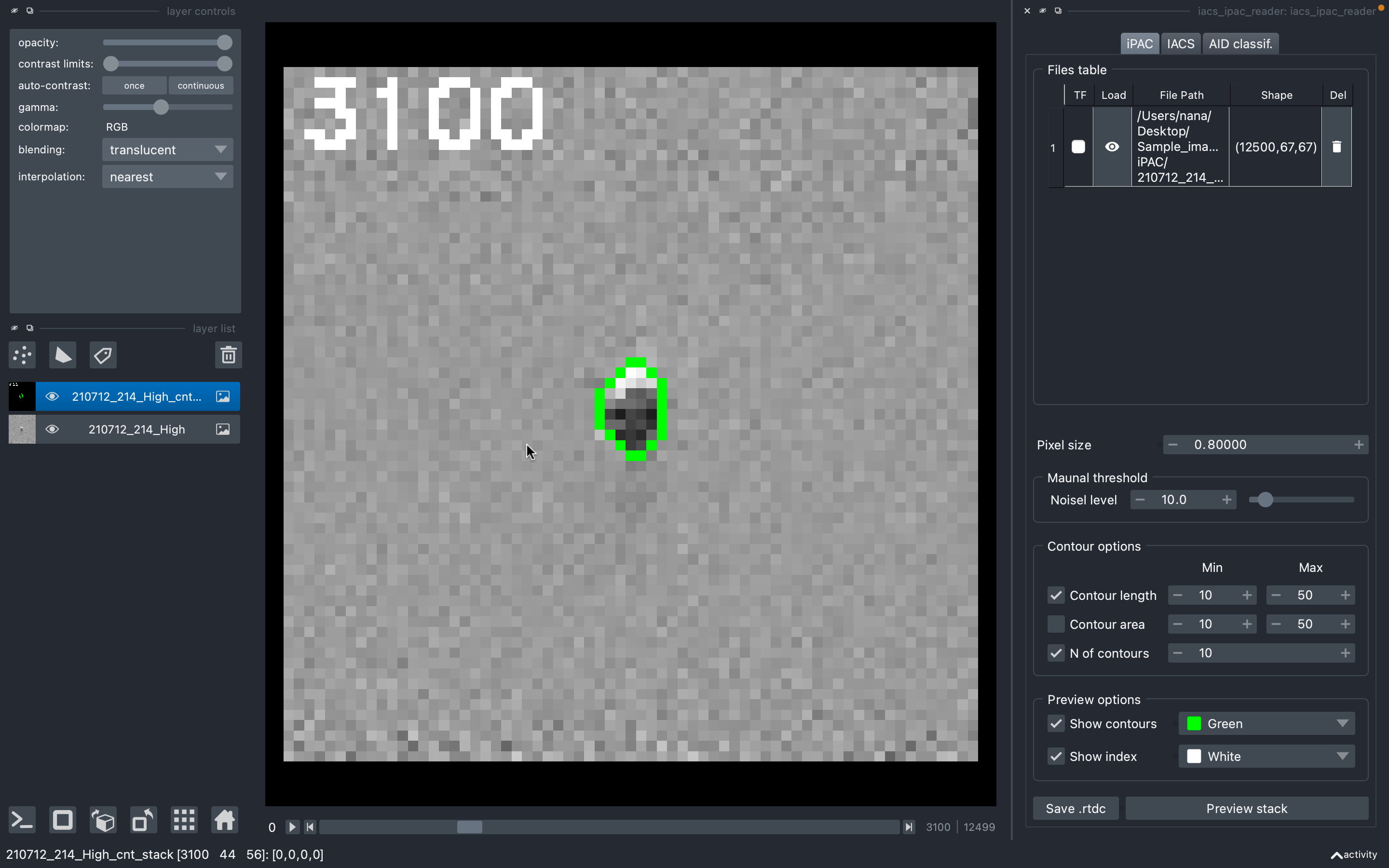
iPAC image contour tracker
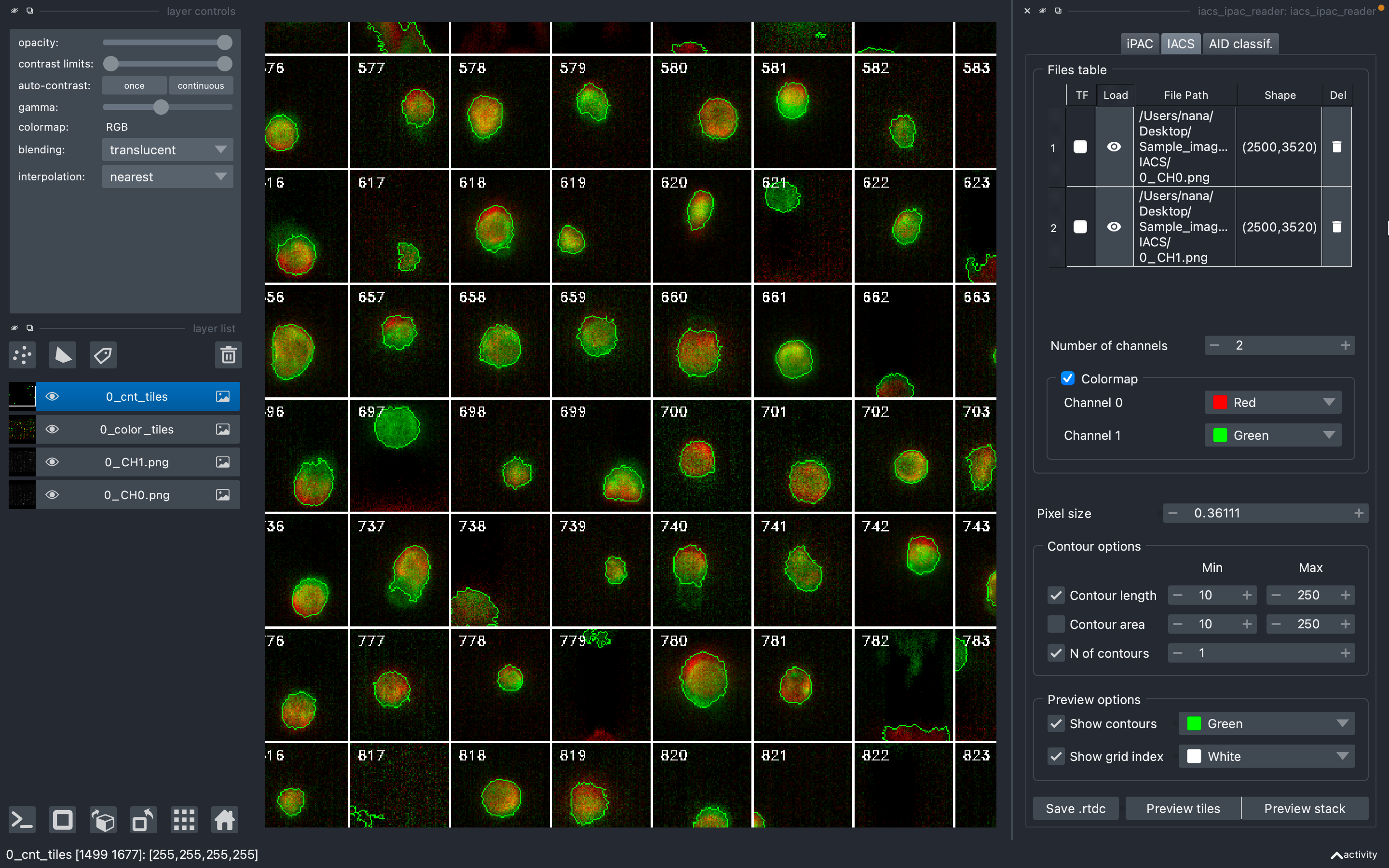
IACS image contour tracker
AID classif.
Contributing
Contributions are very welcome. Tests can be run with tox, please ensure the coverage at least stays the same before you submit a pull request.
License
Distributed under the terms of the BSD-3 license, "iacs_ipac_reader" is free and open source software
Issues
If you encounter any problems, please file an issue along with a detailed description.
Version:
- 0.0.13
Last updated:
- 2022-04-12
First released:
- 2022-01-21
License:
- BSD-3-Clause





