napari-findaureus
Locate bacteria in CLSM obtained infected bone tissue images
"Findaureus" is now available to use in napari.
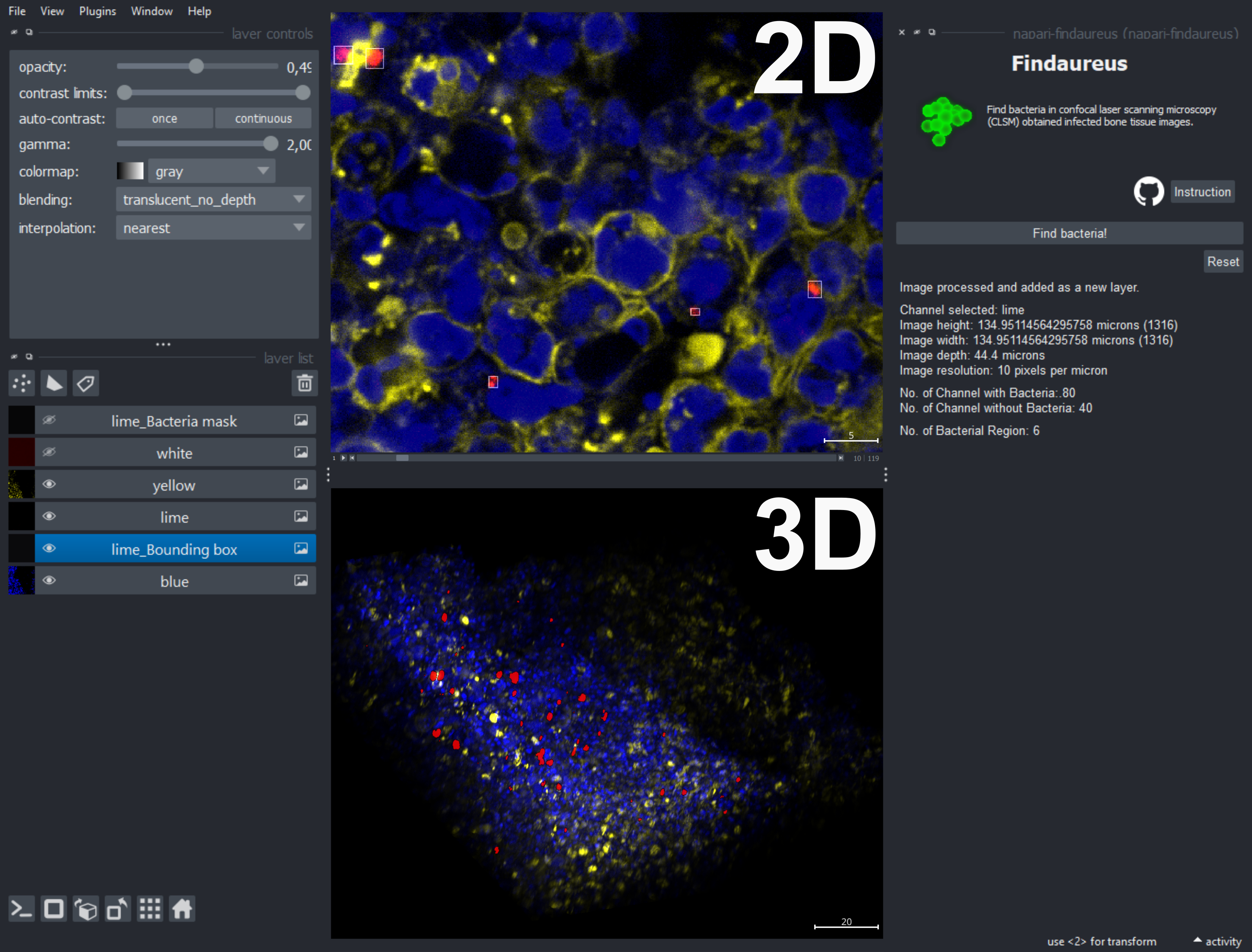
Findaureus is a tool designed to identify bacteria in infected bone tissue images obtained via Confocal Laser Scanning Microscopy (CLSM). This tool can be accessed independently here. Findaureus has been integrated as a plugin for napari. In addition to its bacteria-locating algorithm, the napari viewer provides improved visualization features, in 2D and 3D perspectives.
Installation
Windows/Linux
If you don’t have conda installed, you can get miniconda or Anaconda from their websites.
- Open your command line tool and run these commands to create and activate a conda environment:
conda create -n napari-findaureus python=3.9
conda activate napari-findaureus
- Install napari and napari-findaureus with this command:
pip install "napari[all]" napari-findaureus
macOS
- Create an environment with napari and pyqt5
conda create -n napari-findaureus -c conda-forge python=3.9 pyqt imagecodecs napari
- Install the napari-findaureus plugin
pip install napari-findaureus
Start napari-findaureus
Launch napari from the terminal while the napari-findaureus environment is running.
napari
To launch the napari plugin, go to “Plugins” and select “napari-findaureus”.
Quick demo
To use the napari-findaureus plugin, please follow the steps below:
- First, download some relevant fluorescence-labeled images of infected mouse bone tissues from Zenodo.
- Next, load the image file through the
napari-findaureusplugin. - Navigate to the “Plugins” menu and select the
napari-findaureusoption to activate the widget. - In the viewer, identify the bacteria channel from the "layer list," which is specified in the image file name, and select it.
- Once the bacteria channel is selected, click on the
Find bacteria!button. - The widget will display the image-related data and bacteria count. If you need additional help, click on the
Instructionbutton in the widget. - Before you proceed to another image, reset the viewer by clicking on the
Resetbutton provided in the widget.
Enjoy exploring the fascinating world of bacteria in mouse bone tissues!
Contributing
We welcome and appreciate all contributions to the napari-findaureus project! Whether it's reporting bugs, suggesting new features, improving documentation, or writing code, your involvement is greatly valued.
When using our dataset or referring to our work, we kindly ask that you acknowledge the dataset and cite the related articles. This helps support our work and allows us to continue improving this project.
Thank you for your interest and support!
Citations and Dataset
Findaureus
Mandal S, Tannert A, Löffler B, Neugebauer U, Silva LB (2024) Findaureus: An open-source application for locating Staphylococcus aureus in fluorescence-labelled infected bone tissue slices. PLoS ONE 19(1): e0296854.
Infected mouse bone tissue
Mandal S, Tannert A, Ebert C, Guliev RR, Ozegowski Y, Carvalho L, Wildemann B, Eiserloh S, Coldewey SM, Löffler B, Bastião Silva L, Hoerr V, Tuchscherr L, Neugebauer U. (2023) Insights into S. aureus-Induced Bone Deformation in a Mouse Model of Chronic Osteomyelitis Using Fluorescence and Raman Imaging. International Journal of Molecular Sciences 24(11):9762.
Dataset
Acknowledgements
This project is a part of the European Union's Horizon 2020 research and innovation program under grant agreement No 861122 (ITN IMAGE-IN). We acknowledge support from the Jena Biophotonics and Imaging Laboratory (JBIL), from the European Union via EFRE funds within the Thüringer Innovationszentrum für Medizintechnik-Lösungen (ThIMEDOP, FKZ IZN 2018 0002), the BMBF via the funding program Photonics Research Germany (LPI, FKZ: 13N15713) and via the CSCC (FKZ 01EO1502) and the Institute of Anatomical and Molecular Pathology, University Coimbra, Portugal.
Version:
- 0.0.4
Last updated:
- 2024-04-20
First released:
- 2024-04-20
License:
- MIT
Supported data:
- Information not submitted
Save extension:
Operating system:
- Information not submitted
Requirements:
- magicgui
- qtpy
- napari[all]
- aicsimageio ==4.11.0
- nd2 ==0.5.3
- aicspylibczi ==3.1.2
- fsspec ==2023.5.0
- readlif ==0.6.5
- czifile ==2019.7.2
- tifffile ==2023.7.10
- webcolors ==1.13
- opencv-python ==4.7.0.72
- numpy ==1.24.3
- scikit-image ==0.20.0
- xmltodict
- tox ; extra == 'testing'
- pytest ; extra == 'testing'
- pytest-cov ; extra == 'testing'
- pytest-qt ; extra == 'testing'
- napari ; extra == 'testing'
- pyqt5 ; extra == 'testing'
