DenoiSeg
A napari plugin performing joint denoising and segmentation of microscopy images using DenoiSeg.
A napari plugin performing joint denoising and segmentation of microscopy images using DenoiSeg.
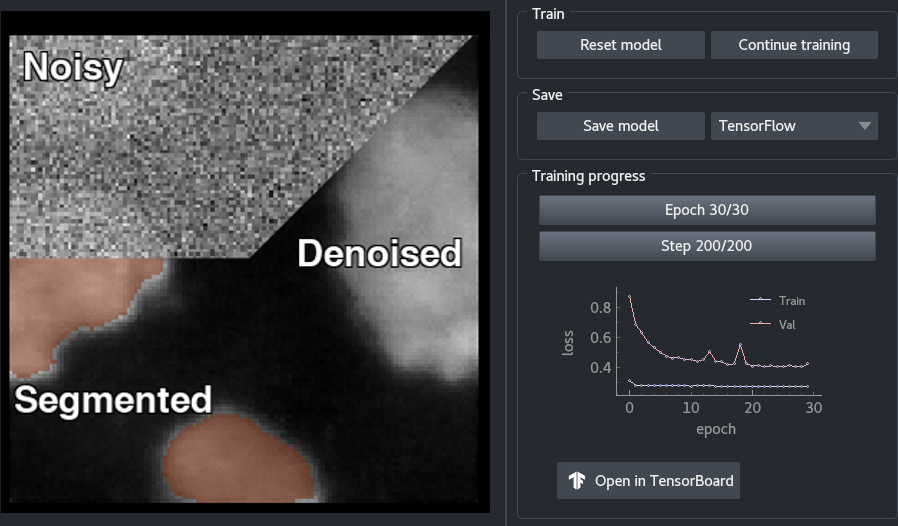 ----------------------------------
----------------------------------
Installation
Check out the documentation for more detailed installation instructions.
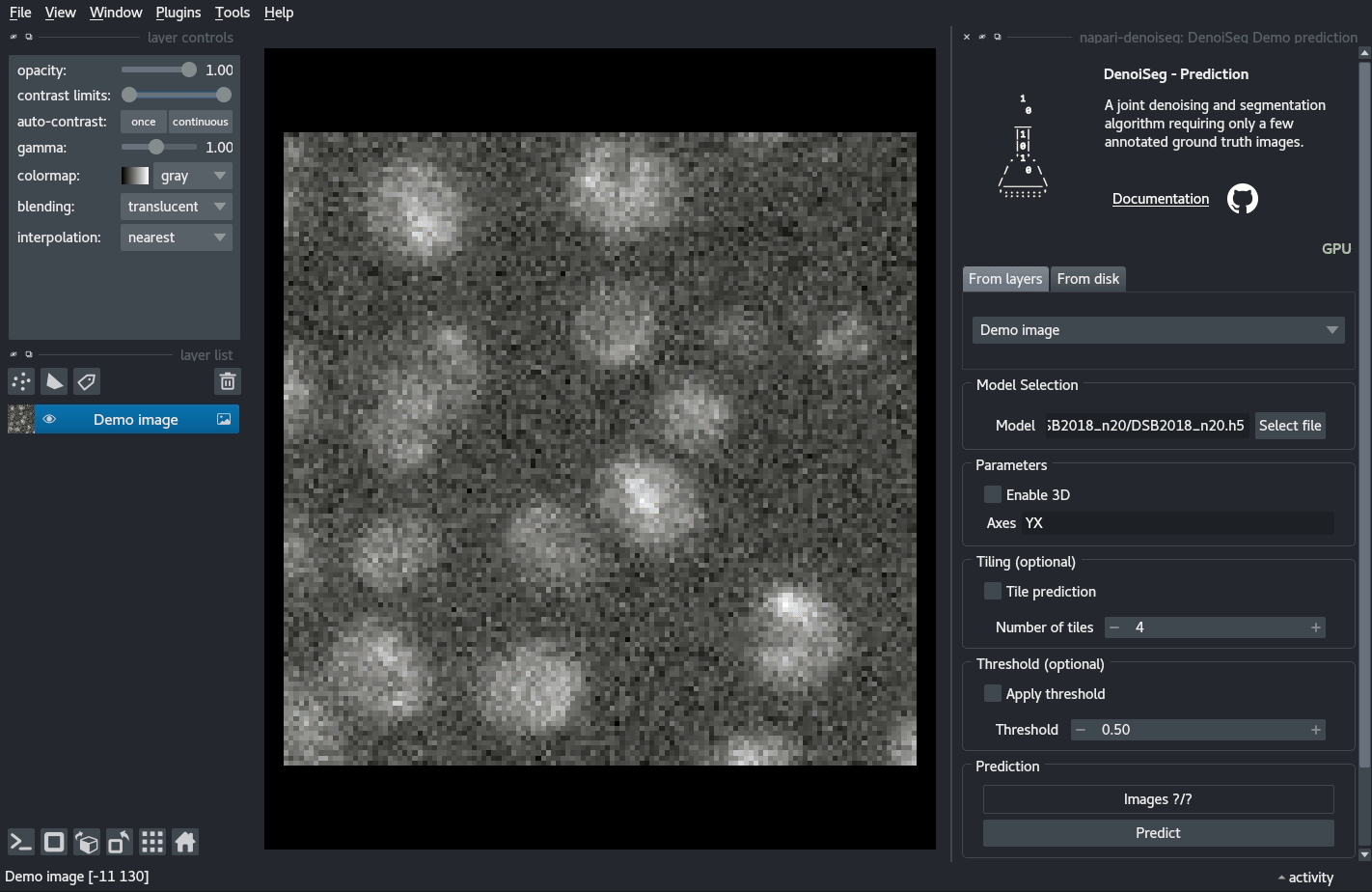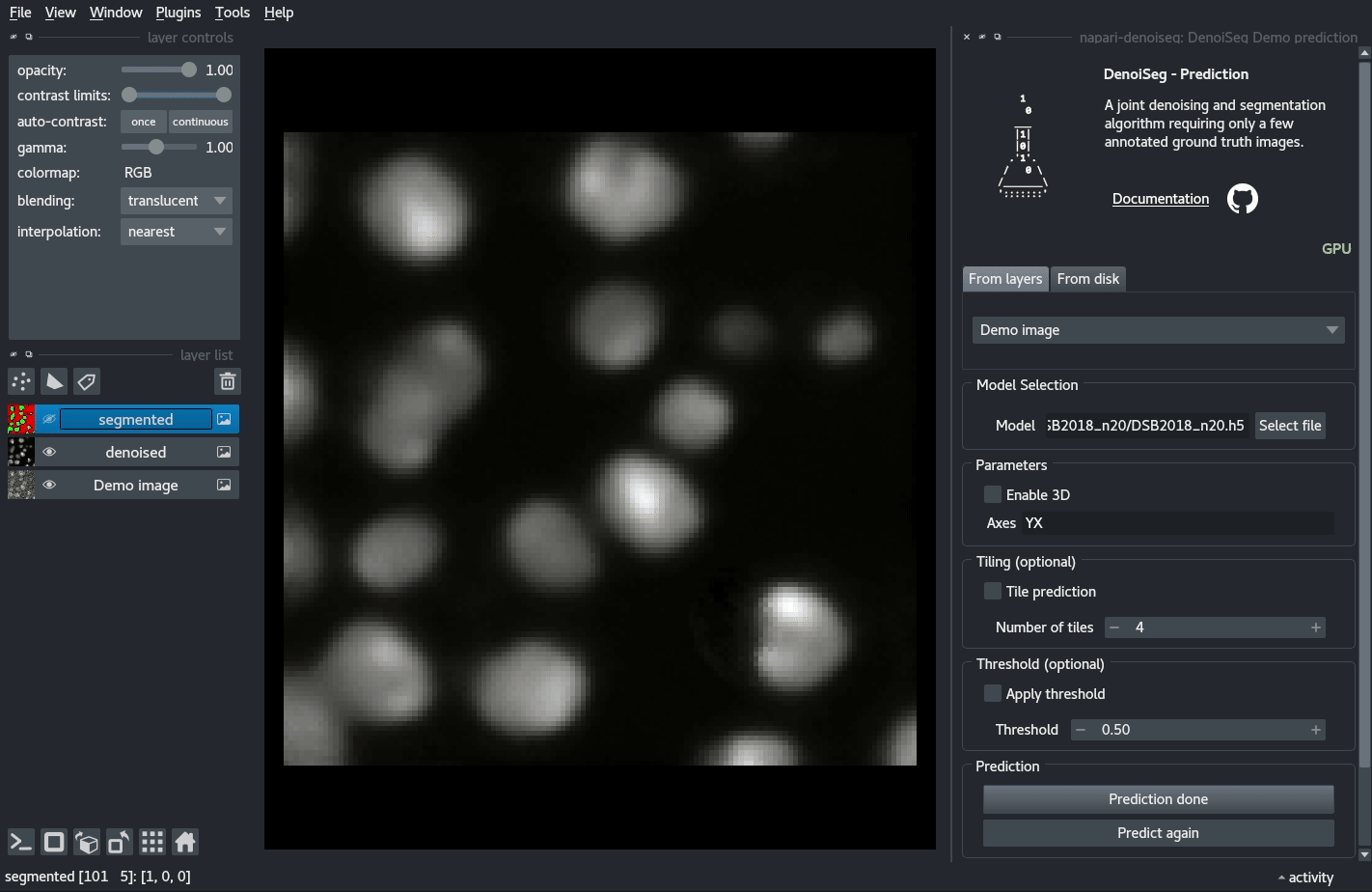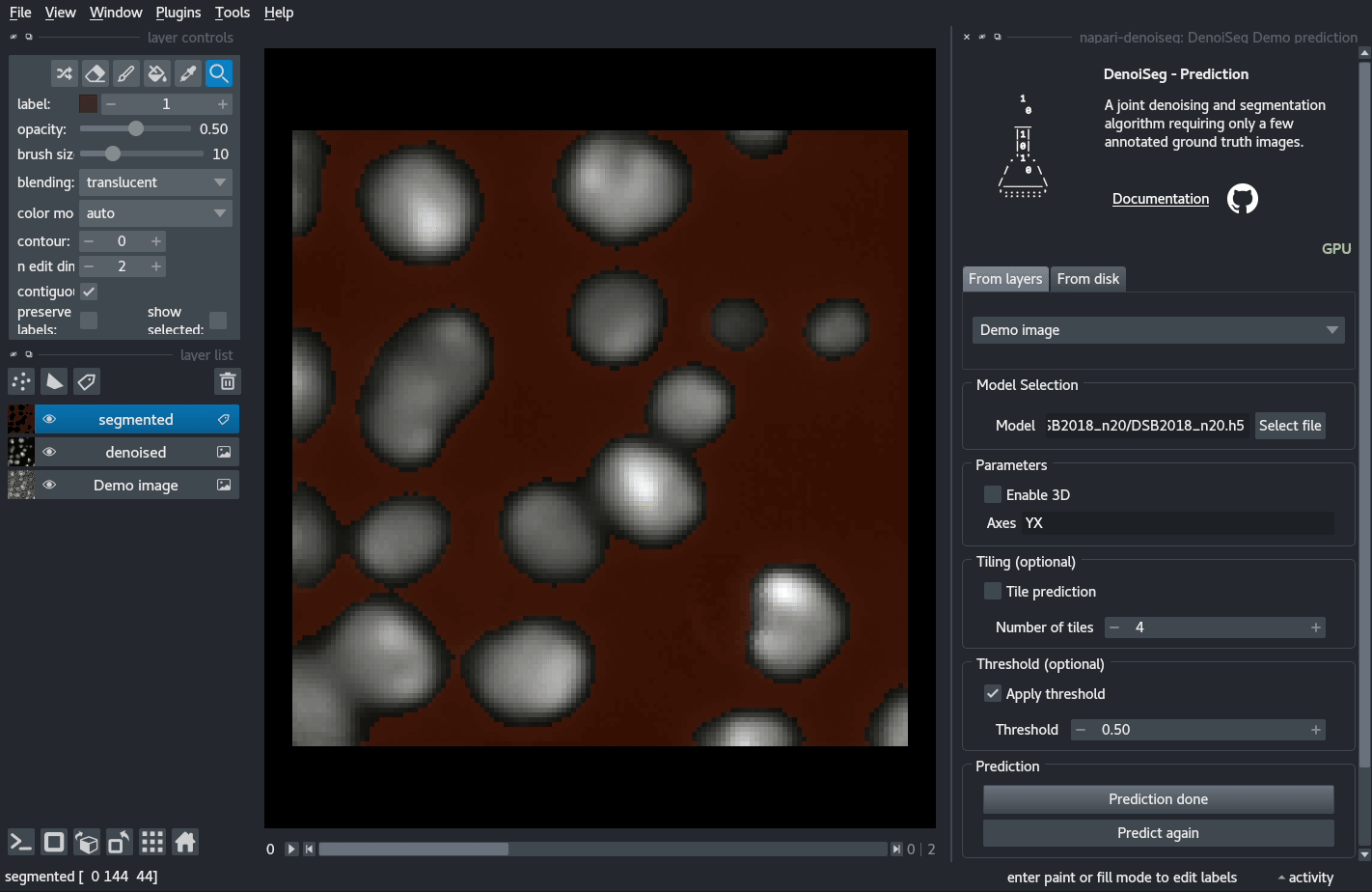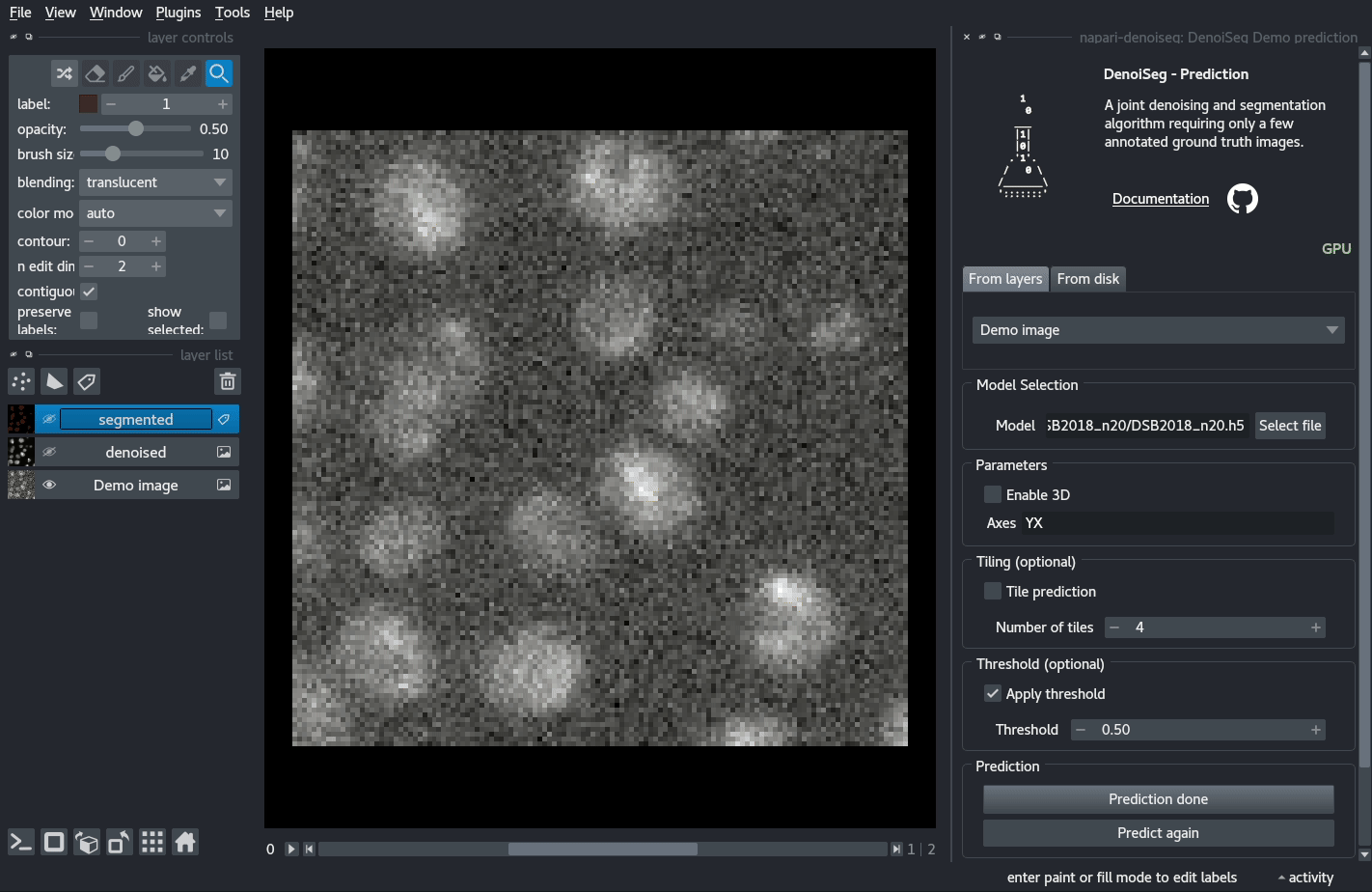
Quick demo
You can try out a demo by loading the DenoiSeg Demo prediction plugin and directly clicking on Predict.
Documentation
Documentation is available on the project website.
Contributing and feedback
Contributions are very welcome. Tests can be run with tox, please ensure the coverage at least stays the same before you submit a pull request. You can also help us improve by filing an issue along with a detailed description or contact us through the image.sc forum (tag @jdeschamps).
Cite us
Tim-Oliver Buchholz, Mangal Prakash, Alexander Krull and Florian Jug, "DenoiSeg: Joint Denoising and Segmentation" arxiv (2020)
Acknowledgements
This plugin was developed thanks to the support of the Silicon Valley Community Foundation (SCVF) and the Chan-Zuckerberg Initiative (CZI) with the napari Plugin Accelerator grant 2021-239867.
Distributed under the terms of the BSD-3 license, "napari-denoiseg" is a free and open source software.
Version:
- 0.0.1rc2
Last updated:
- 2022-11-01
First released:
- 2022-10-31
License:
- BSD-3-Clause
Supported data:
- Information not submitted
Plugin type:
Open extension:
Save extension:
Operating system:
- Information not submitted
Requirements:
- numpy
- pyqtgraph
- denoiseg (>=0.3.0)
- bioimageio.core
- magicgui
- qtpy
- napari-time-slicer (>=0.4.9)
- napari (<=0.4.15)
- vispy (<=0.9.6)
- imageio (!=2.11.0,!=2.22.1,>=2.5.0)
- tensorflow ; platform_system != "Darwin" or platform_machine != "arm64"
- tensorflow-macos ; platform_system == "Darwin" and platform_machine == "arm64"
- tensorflow-metal ; platform_system == "Darwin" and platform_machine == "arm64"
- pytest ; extra == 'testing'
- pytest-cov ; extra == 'testing'
- pytest-qt ; extra == 'testing'
- pyqt5 ; extra == 'testing'





